Blog
The holobiont paradigm finally acknowledges that the development and health of macroorganisms, as all other natural ecosystems, are shaped by the complex microbial communities they host. Yet, mechanistic insights into microbial community assembly and their interactions with their environment are still scarce. While it is currently estimated that less than 1% of prokaryotic species can be cultivated in the lab, meta-omics (especially metagenomics) have been the top microbiology methods over the past 2 decades. They allowed a significant number of high-impact discoveries, but they also raised an even more significant number of questions. Be it for fundamental research purposes or with a view to provide reasoned solutions, there is now a need for stepping back from empirical approaches and gaining knowledge of the mechanisms underlying the observed patterns [1].
Let’s take an example: various woman (and infant!) health conditions, be it immunity, fertility, or cancer risks, are associated to different vaginal “microbial signatures”, but in which direction does this association go? Can we tell for sure that vaginal dysbiosis is the reason for endometriosis, does that work the other way around, or are they both coincidental consequences of something else, like contraception or nutrition? And from there, do we have the means to manipulate the microbiota to sustainably restore health conditions?
A knowledge upshot is expected thanks to the development of single microbial cell approaches, as it will:
- Enable single microbial cell omics, that is characterization of populations and subpopulations, as well as insights into community assembly processes.
- Help crossing the barriers of cultivability, and accelerate the discovery of new microbial physiologies, by boosting the throughput of culture library generation.
We will take you here through both of these facets.
Looking at single microbial individuals to understand microbial communities, a paradox?
It may seem paradoxical that getting knowledge of single individual cells can shed light on community and ecosystem functioning. Still, single microbial cell -omics will significantly contribute to microbial ecology and microbiology in general as (i) it gives access to population and subpopulation diversity and evolutionary patterns, and (ii) it tackles some of the main weaknesses of meta-omics, (iii) offering a complementary perspective of the same object, while (iv) allowing finer and more accurate view of physiological networks [2]. Single cell -omics applied to microorganisms also come with major challenges, that will be described and discussed here.
Microbial interactions, from predation to symbiosis, through competition and mutualism, occur at the individual, population, community and environment levels (Figure 1). Those are intricated and interconnected ecological scales. Meta-omics techniques span from community to environmental scales in microbial ecology but lack the resolution to capture finer scales this ecological gradient, that are population and subpopulation scales. Microbial (sub)populations, e.g., species and lower taxonomic ranks, represent an understudied reservoir of genotypic and phenotypic diversity, and accessing the intra-populational level allow fine scale analysis of evolutionary processes, gene acquisition/loss processes, population emergence and, ultimately, community assembly. Used appropriately, especially in terms of sampling strategy and sample size, single-cell omics encompass the full ecological continuum, from the individual to the community in the same sample, offering the opportunity to connect information from all those levels.
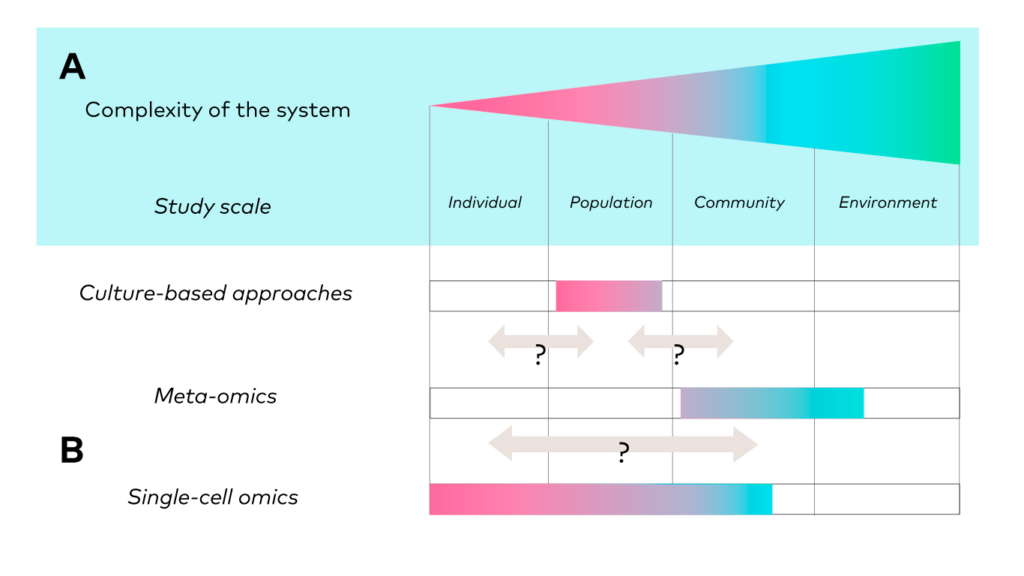
Figure 1. Gradient of ecological scales and associated approaches. From Mauger et al. 2021, Trends in Ecology & Evolution [3]
Meta-omics outputs an averaged information, across a sample of available molecular information within an environment. Drawing population-level insights from meta-omics data remains challenging: during the process of genome reconstruction (metagenomics assembled genomes, or MAGs), the joining of fragments from different individual genomes and/or free DNA can lead to the formation of chimeras (Figure 2). The accuracy of MAGs becomes a formidable challenge when microbial communities are very diverse in populations, with closely related taxa sharing highly similar genomes. In addition, metagenomics does not discriminate DNA contained in living cells from free DNA, that may persist for long periods in the environment, or DNA contained in dead cells [4]. Therefore, it remains impossible to directly link a detected gene to their original microbial cell, nor to the other genes it contains, from meta-omics data. Single-cell omics, rapidly gaining popularity in microbiology, address this issue, by completely shifting the perspective. They focus on the DNA, RNA or proteins contained in the very same cell, opening new possibilities for studying metabolic pathways or gene networks. Recent advances in the field showed that the observation of microbial communities from metagenomics and single cell whole genome sequencing (scWGS) give different representation of the diversity within the same samples [5]. It suggests that the most comprehensive approach to microbial community composition and function could be to apply both meta-omics and single-cell omics to the same sample. This combines the advantages of both techniques: (i) the ability to analyze alpha/beta diversity with high throughput with metagenomics, and (ii) the capacity for detailed and accurate analysis through scWGS.
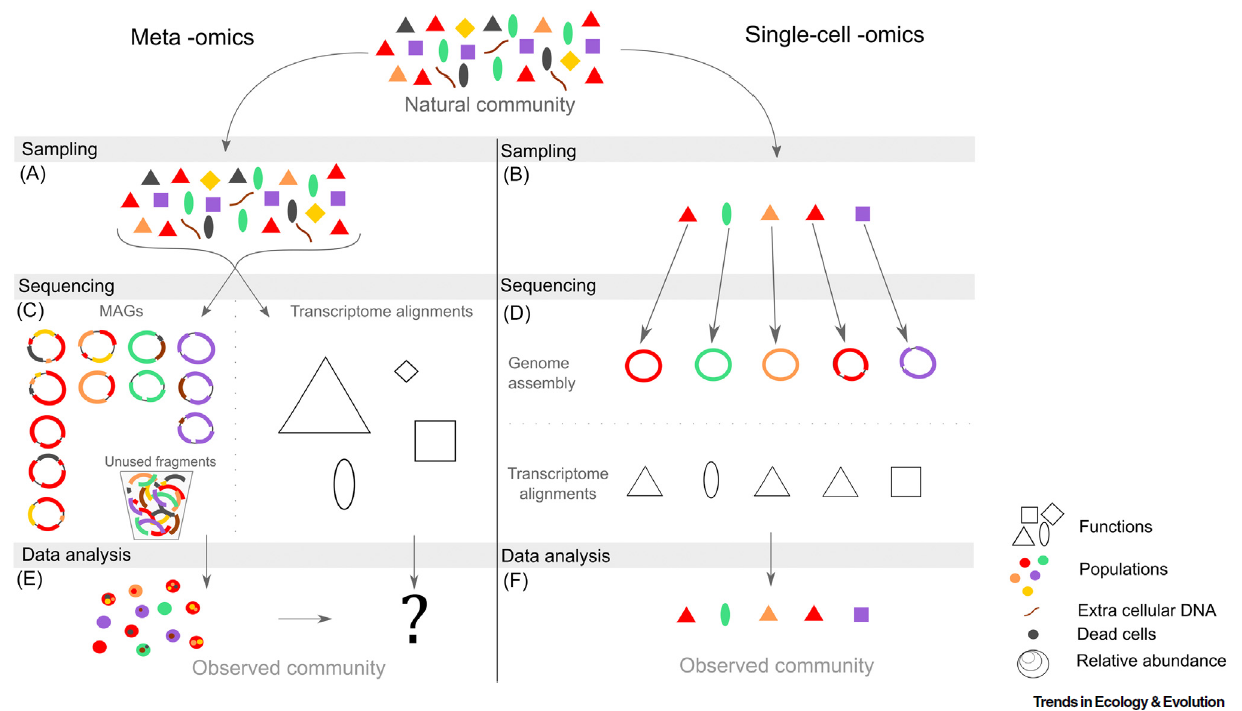
Figure 2. The key steps, features and limitations of meta -omics (left) and single-cell -omics (right). From Mauger et al. 2021, Trends in Ecology & Evolution [6]
Single-cell omics applied to microbes come along with important challenges:
- Cells should be isolated, i.e., detected and separated, accurately and monoclonality status (i.e., one and only one cell) should be verifiable. Most isolation methods (including FACS) require fluorescence staining to accurately detect and successfully isolate (single) small cells, which comprises important biases and is time- and cost-consuming.
- To avoid the loss of the cell or its molecular material, cells need to be deposited in compartments compatible with molecular biology workflows, and subsequent reactions should be compatible with each other as no purification step is possible before cell barcoding (see below).
- Cell material (DNA, RNA or other molecules of interest) is present in very small quantities, especially for bacterial and archaeal cells, which contain just a few femtograms of DNA and RNA (about 1000-fold less than human cells). DNA and cDNA must then be amplified for preparation of sequencing libraries for scWGS and scRNAseq, respectively. Any contamination by foreign DNA (e.g., host DNA), which would be amplified together with the cell material, may then dramatically spoil the data. [7]
- Sequencing costs are still high, and library preparation reagents are a big part of the total experimental costs.
We argue that cellenONE is the optimal solution for single-cell omics of microbial cells. It is the combination of high-precision nanolitre-volume liquid dispensing and image-based cell isolation:
- It can isolate any type of cells from 0.5 to 80 µm, whether microbial, animal or plant, with high accuracy (up to 100% for bacteria).
- It can deposit these cells one by one, in any type of consumables, standard like multi-well plates, or customised.
- For image-based isolation, the user selects cells of interest by setting parameter ranges (size, morphology, fluorescence), and microscopy images are recorded for all isolated cells, allowing QC. They can therefore target specific types of cells, for instance only prokaryotes and exclude host cells.
- While possible, fluorescence staining is not necessary: cellenONE reaches maximum accuracy for single cell isolation even in bright field illumination, when using its proprietary machine-learning microLIFE software for small cells (available since January 2024).
- The instrument exists as a version embedded in a biosafety cabinet (cellenONE BSC), and its capillary is inert and sterilizable, to maintain axenic conditions and avoid contamination.
- It generates ultra small droplets (100-500 pL) so that the carry-over liquid in the droplet containing the cells, hence the probability of free DNA contamination, is very limited.
- It can be used as a liquid dispenser to generate miniaturized molecular biology reagents for drastic cost reduction.
Single-cell -omics may also be a formidable way to learn more about microbial physiology, by accessing gene assemblage and pathway networks at the individual cell level. It was proven efficient, for instance, at providing data allowing to infer metabolic preferences, then used for choosing appropriate enrichment conditions, leading to successful cultivation, pushing the borders of microbial dark matter [8].
Bring ‘microbial dark matter’ to light by cultivating the non-cultivable.
Microbial dark matter refers to the fraction of microbes that we are not able to cultivate, by lack of knowledge or control of their growth requirements. It is generally admitted that more than 99% of microbes (especially bacteria and archaea) are non-cultivable in the lab, especially when they belong to less understood (because less studied) ecosystems such as soils [9]. Molecular studies confirm the prevalence of taxa detected through their genetic elements with little to no culture representatives, even many ubiquitous and abundant ones like Acidobacteria in soils [10]. As a result, microbial physiology studies critically focus on a very small portion of living microorganisms, and extrapolation of culture-based approaches findings to natural communities is risky, if not fictional.
What if we could cut down the share of microbial dark matter? Let’s start over from the first sentence of this section and rephrase it: microbial dark matter refers to the fraction of microbes that we are not YET able to cultivate. We argue that (i) increasing the throughput of culture generation is the key to ‘microbial brightened matter’ and that (ii) it will be unlocked by automated single microbial cell isolation.
Conventionally, the most used methods for culture generation are:

- Direct plating (Figure 3A): a microbe suspension solution is serial diluted then plated on agar medium. After a first incubation, colonies are picked (manually or with a colony picker) and placed in liquid medium. After a second incubation, the liquid cultures are most often submitted to at least one streaking step on agar plate, to guarantee the purity of the isolated strain. After a third incubation, streaked colonies are picked again and placed in liquid medium so that, after a fourth incubation, cultures can be stored and/or used for downstream analysis.
- Dilution to extinction (Figure 3B): a microbe suspension solution is serial diluted then inoculated in liquid cultures (generally in microplates). This method relies on Poisson law, e.g., on the chance that one well is inoculated by one single cell. After a first incubation, the plates showing a proportion of wells statistically consistent with a high proportion of single-cell inoculation are chosen, and each of the grown wells is streaked for purification, at least once. After a second incubation, streaked colonies are picked again and placed in liquid medium so that, after a fourth incubation, cultures can be stored and/or used for downstream analysis.
This has severe limitations, that restricts culturomics outcomes. First, it is far more boring and long than reading the protocol description (!). The generation of culture collections is tedious and time-consuming: it occupies all of your space, all of your consumables, all of your time. This limits the throughput, hence the depth of the approach, while it is shown that the multiplication of growth conditions (nutrient availability, incubation temperature, oxidoreduction conditions, and combinations of those), is crucial for isolation of diverse libraries [11] [12]. Second, mixed colonies can form, featuring cells belonging to two or more microbial populations, species, genera or even kingdoms. Obtaining monoclonality (that is a culture starting from a single clone) can be difficult if not impossible to achieve through direct plating or dilution to extinction, even when repeating streaking steps. Third, some microbes may simply not be able to grow on agar media (many archaea taxa for instance), therefore streaking them is impossible.
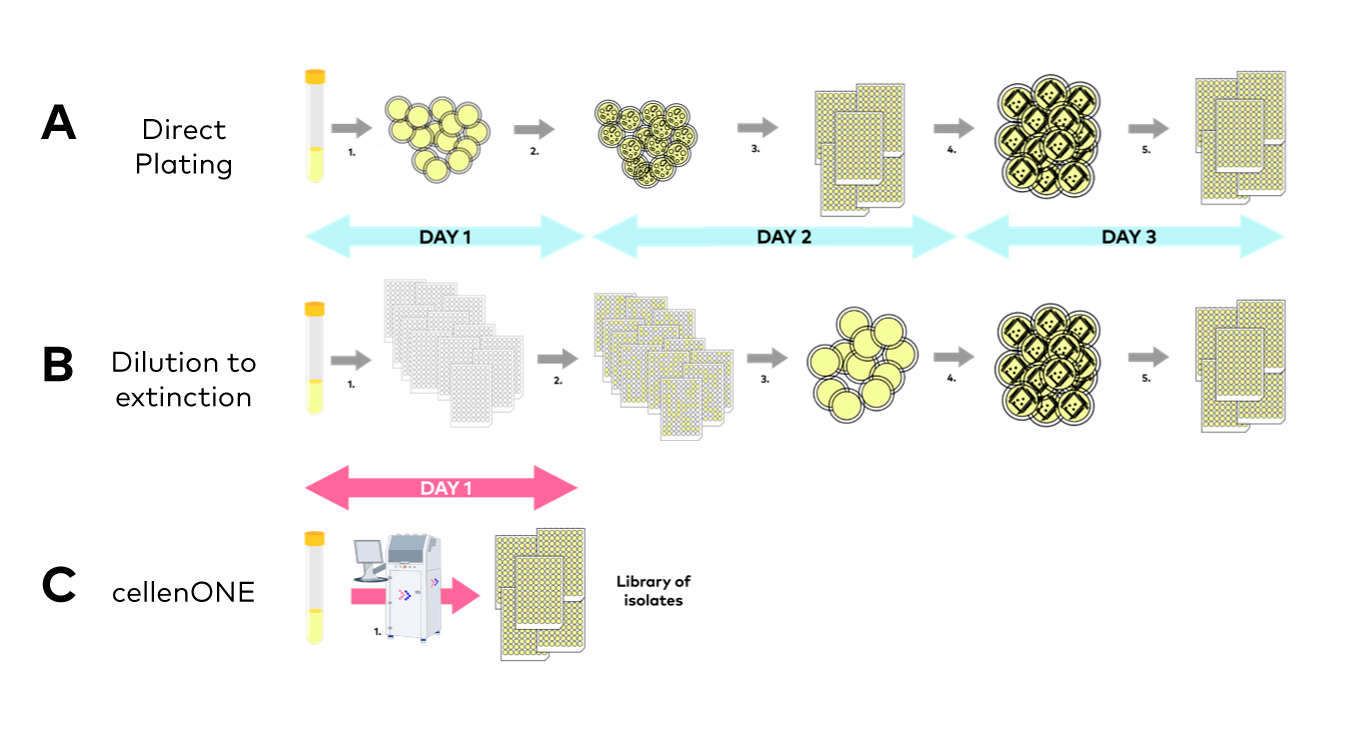
Figure 3. Classical approaches to culture library generation, i.e., Direct plating (A), and Dilution to extinction (B) vs. single microbial cell inoculation of cultures by cellenONE (C).
Single cell isolation automation using cellenONE can help alleviating all these limitations:
- With demonstrated innocuity on microbial growth, cellenONE can isolate microbial cells in any type of consumables, filled with any type of medium, liquid or solid.
- Because microscopy images are taken and recorded for all isolated cells, monoclonality can be checked live on the instrument’s screen, and streaking steps become unnecessary, considerably shortening both hands-on and total experimental time (Figure 3C).
- If the microbial suspension comes from a mixed enrichment or colonies, cells can be separated based on morphological criteria.
- The instrument is automated and can inoculate cultures with single clones at high throughput, opening fast and efficient one-to-many and many-to-one approaches [13].
Altogether, the technological breakthrough allowing accurate single microbial cell approaches constitutes a formidable opportunity for all fields of microbiology. This represents a shift of perspective, from satellite view to macroscopic lens, that will unlock mechanistic insights into microbiota assembly and physiology.

References and notes:
[1] Prosser, J. I. (2015). Dispersing misconceptions and identifying opportunities for the use of’omics’ in soil microbial ecology. Nature Reviews Microbiology, 13(7), 439-446.
[2] Blainey PC. (2013). The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev, 37(3), 407-27.
[3] Mauger, S., Monard, C., Thion, C., & Vandenkoornhuyse, P. (2022). Contribution of single-cell omics to microbial ecology. Trends in Ecology & Evolution, 37(1), 67-78.
[4] Persistence of free material is arguably less of an issue for metatranscriptomics and metaproteomics, as RNA and proteins have substantially shorter half-life time than DNA.
[5] Mauger, S., under the direction of Vandenkoornhuyse, P. & Monard, C. (2023). Elaboration of an innovative single-cell genomics approach and application to soil bacterial communities. Doctoral dissertation, Université de Rennes.
[6] Mauger, S., Monard, C., Thion, C., & Vandenkoornhuyse, P. (2022). Contribution of single-cell omics to microbial ecology. Trends in Ecology & Evolution, 37(1), 67-78.
[7] For single cell proteomics (scMS), no amplification is possible so that the feasibility of scMS applied to microbial cells rely on mass spectrometer sensitivity, which is constantly increasing these last years.
[8] Weber, E., under the direction of Gubry-Rangin, C. & Prosser, J. (2016). Ecological insights into unexplored Archaea through environmental ecophysiology, single-cell genomics and cultivation. Doctoral dissertation, University of Aberdeen.
[9] Pham, V. H., & Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends in biotechnology, 30(9), 475-484.
[10] Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., & Suriani, N. L. (2020). Recent understanding of soil acidobacteria and their ecological significance: a critical review. Frontiers in Microbiology, 11, 580024.
[11] Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., … & Raoult, D. (2018). Culturing the human microbiota and culturomics. Nature Reviews Microbiology, 16(9), 540-550.
[12] Vanstokstraeten, R., Mackens, S., Callewaert, E., Blotwijk, S., Emmerechts, K., Crombé, F., … & Demuyser, T. (2022). Culturomics to investigate the endometrial microbiome: proof-of-concept. International Journal of Molecular Sciences, 23(20), 12212.
[13] For instance: one sample to many different growth conditions (media, temperature etc.), each of the many cells of a sample to wells containing a compound of interest for high throughput metabolic screening etc.
