Single Microbial
Cell Isolation
Access the microbial individual level
Single Microbial
Cell Isolation
Access the microbial individual level
cellenONE is the optimal tool for sorting and isolation of microorganisms for both single cell omics analyses and culture-based applications
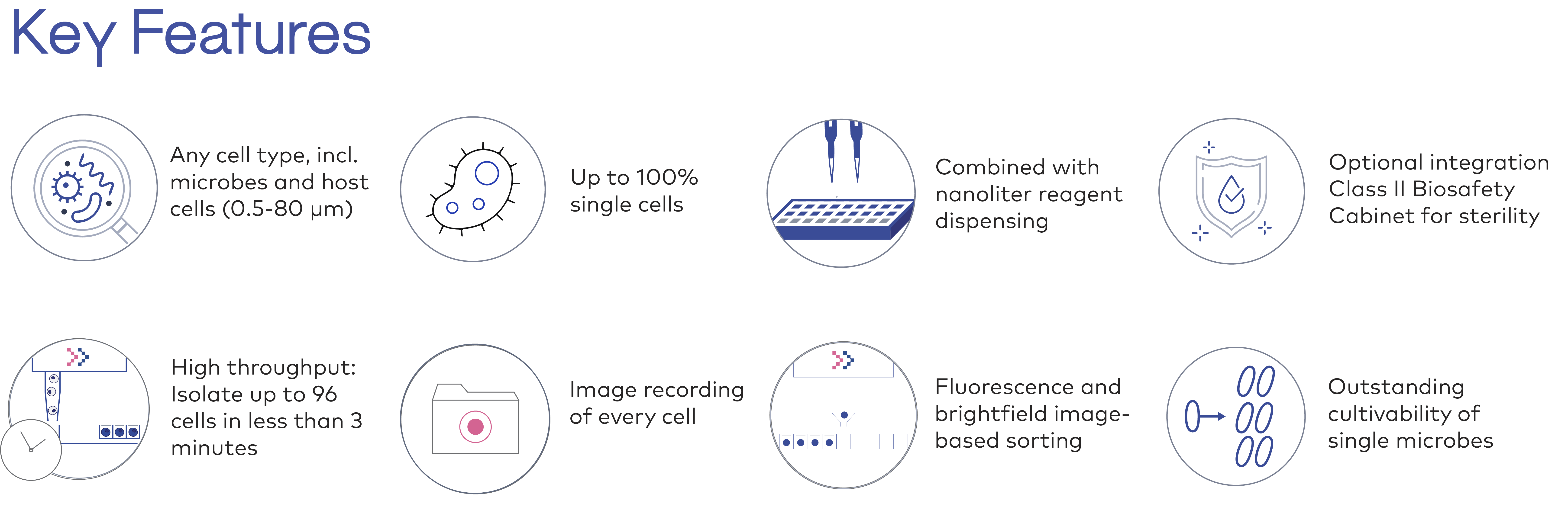
A unique cell type range: any microbe, any size.
Here is what we have tried so far, and we will be very happy to try other microorganisms that you would like to isolate!
Bacteria and archaea, for instance:
- Bacillus
- Escherichia
- Lactobacillus
- Microccocus
- Pseudomonas
- Staphylococcus
- Haloferax
Filamentous fungi spores and conidia, and yeasts, for instance:
- Aspergillus Niger
- Aspergillus Oryzae
- Saccharomyces cerevisiae
- Botrytis cinerea
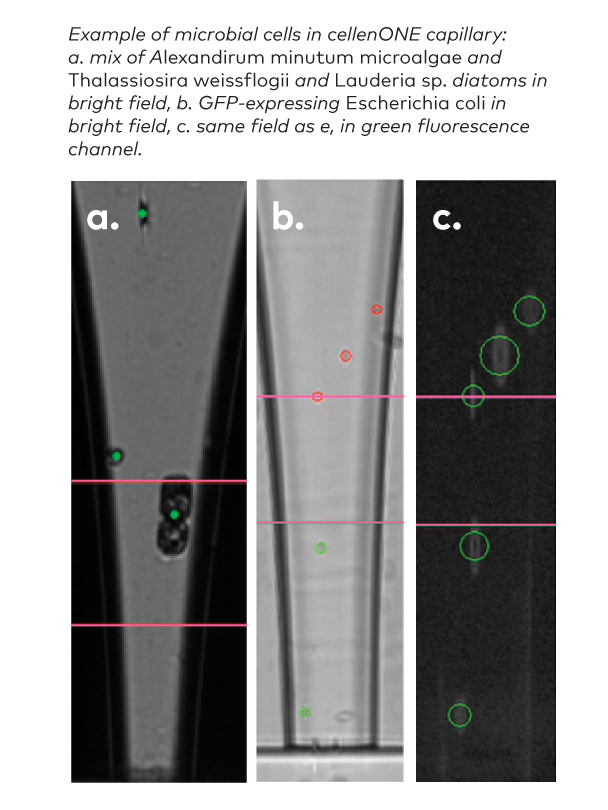
Autofluorescent or not, for instance:
- Alexandrium Minutum
- Thalasiossira Weissflogii
- Lauderia sp.
- Dictyostelium
- Leishmania
The optimal tool for clone isolation in synthetic biology
Synthetic biology is the new chemistry: bioengineered microbes can be used to produce any kind biomolecules, from drugs to fragrances.
- cellenONE can dispense single clones directly in liquid cultures, scaling up mutant screening by automated pure culture generation.
- Combine high performance on bacteria, fungal spores, yeasts and microalgae, easy sample prep and traceable post-isolation QC.
- cellenONE goes HT, for integration in your production process and Cellenion engineering team helps you for an optimal and fast set-up of your process.
An open platform for microbial single cell omics
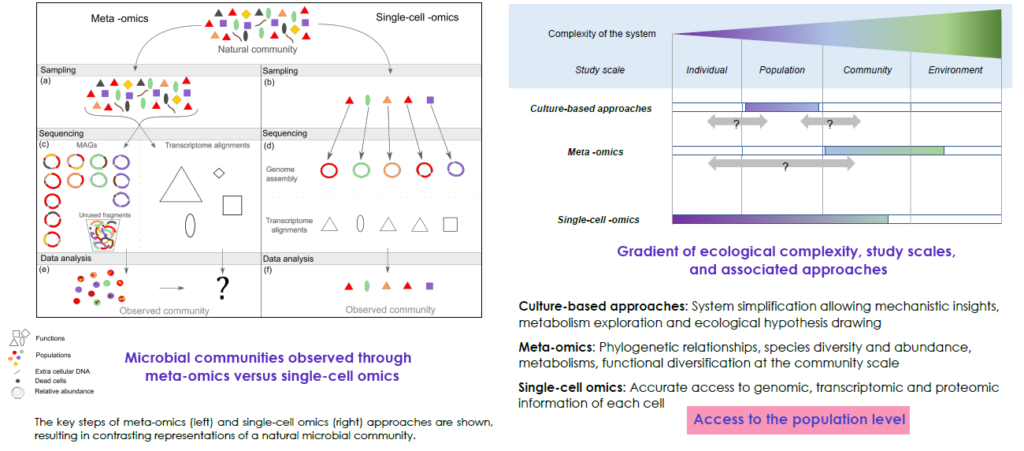
Metagenomics and metatranscriptomics studies led to significant discoveries in microbiology, but they also gave rise to many more theories and hypotheses. While meta-omics provide averaged information, rarely disentangled, a need emerges to access the individual microbial cell level.
- cellenONE is compatible with virtually any single cell -omics methods (single cell whole genome sequencing, RNA sequencing and proteomics) and can be combined with cutting edge –omics technologies (e.g., digital PCR).
- Reduce cost and contamination in account of cellenONE nano-volume liquid dispensing ability.
Cellenion is collaborating with the University of Rennes and CNRS to develop single cell –omics solutions for microbial cells, within the framework of the funded project LabCom MICROSCALE.
Cellenion and ECOBIO laboratory (UMR CNRS-University of Rennes, expert in microbial ecology and environmental genomics) received a Labcom-2020 €360k grant by the French National Research Agency (ANR) for the creation of the joint MICROSCALE-Lab (MICRObial Single Cell AnaLysEs Laboratory). The overall objective of this 5-year project is to offer a complete solution for the isolation, library preparation and bioinformatic analysis of single microbial cells, creating a conceptual and technological breakthrough for the study of microbial communities. A knowledge upshot is expected in microbial interactions and eco-evolutionary boundaries through the enabling of mechanistic characterization of populations and community assembly processes. Currently, the use of meta-omics and single-cell omics on the same
sample appears to be the best option, combining the strengths of the two approaches: (i) high throughput and alpha/beta diversity and (ii) fine-scale analysis by scWGS and/or scRNAseq.
Labcom latest news:
Solène Mauger, joint student of Ecobio lab and Cellenion, successfully defended her PhD thesis “Elaboration of innovative single-cell genomics approach and application to soil bacterial communities”. She will start in February 2024 a Post-doc at Ecogeno, dedicated to the development of microbial scRNAseq workflow.
Earlier in the project:
Check out our publication on the contribution of single microbe -omics to microbiology:
High throughput culturomics
Culture-based approaches are rising back in microbiology. Classical methods for isolate library generation are tedious and time-consuming, therefore increase in throughput is now needed.
- cellenONE can isolate any type of cells, and mixes of those types, including host and microbial cells in the same sample!
- With no need for fluorescent staining, cellenONE ensures optimal post-isolation cultivability, for abundant and diverse culture libraries.
Unique single cell accuracy
- Best-in-class single cell accuracy for all types of cells
- With our microLIFE software, cellenONE is now able to reach up to 100% single cells for all types of microbes (even the smallest ones), both in bright field and fluorescence: no equivalent on the market.
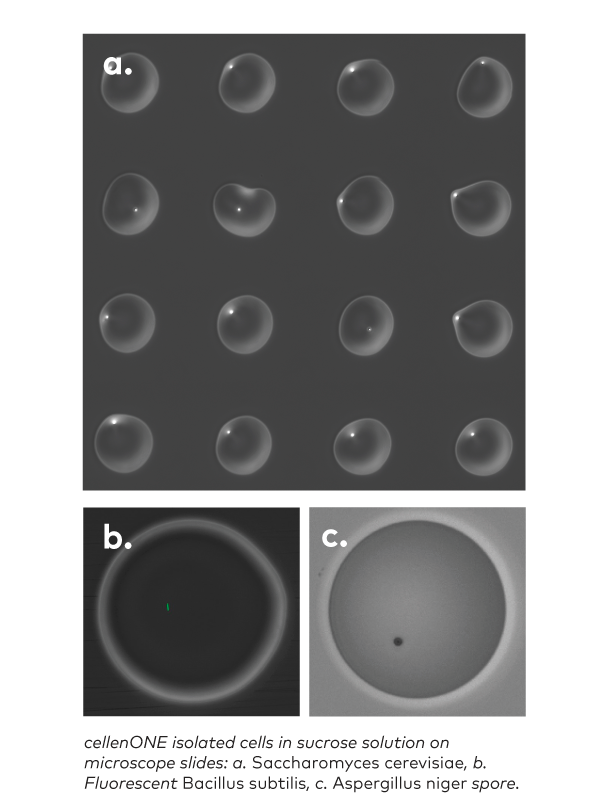
Image-based cell isolation
- User-defined sorting parameters (morphology and/or fluorescence)
- 4-channel fluorescence (DAPI, FITC Cy3, Cy5)
- Assurance of monoclonality: recorded images
- Immediate visual feedback on sample preparation (e.g. presence of host cells, debris, etc…)
- Link phenotypic and morphological information with omics data
Use any target labware
- Microorganisms can be isolated onto or into any consumable, MTP, petri dish, microscope slide, etc.
- Very high positional precision of cell isolation and reagent dispensing onto custom chips and microdevices
Nanoliter liquid dispensing for reaction miniaturization
- Small droplet volume (150 to 600 pL)
- Compatible with a wide range of reagents (incl. culture media, molecular biology reagents…)
- Temperature and humidity control to prevent evaporation and enable on-deck incubations
