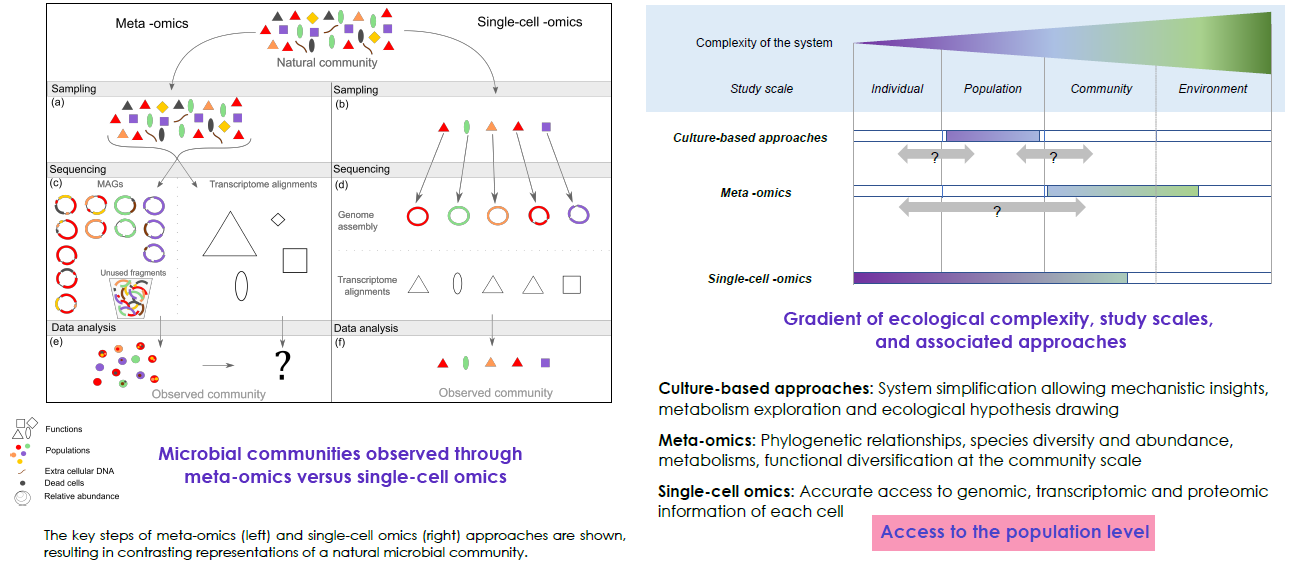
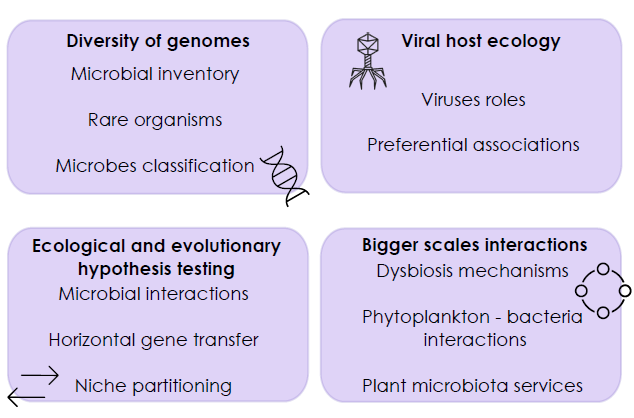
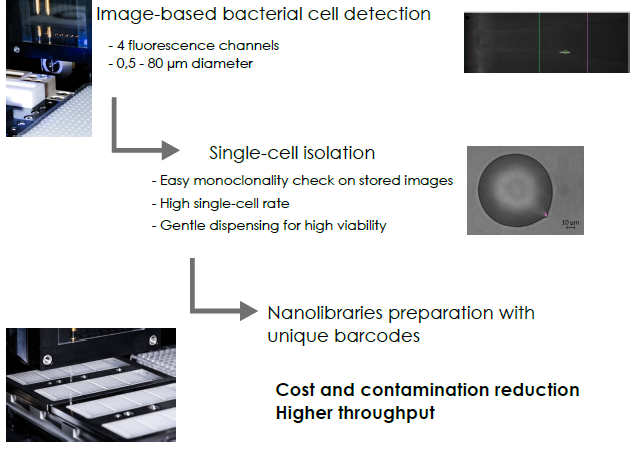
Single-cell omics applied to micro-organisms could become a gold standard in microbial ecology. A knowledge upshot is
expected in microbial interactions and eco-evolutionary boundaries through the enabling of mechanistic characterization
of populations and community assembly processes. Currently, the use of meta-omics and single-cell omics on the same
sample appears to be the best option, combining the strengths of the two approaches: (i) high throughput and alpha/beta
diversity and (ii) fine-scale analysis by scWGS and/or scRNAseq.
This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.