Single Cell Proteomics
The Promise of Single-Cell Proteomics for Biology and Medicine
Single-cell proteomics (SCP) is an emerging field that uses mass spectrometry to measure thousands of proteins in individual cells. Unlike single-cell transcriptomics, which reflects potential activity, SCP reveals the actual molecules driving cell function, signalling, and disease mechanisms. This makes it a powerful complement for precision medicine, helping researchers identify rare pathogenic cells, track drug responses at single-cell resolution, and uncover new therapeutic targets.
The recent leap in mass spectrometry (MS) sensitivity, driven by nanoscale liquid chromatography MS (LC-MS/MS), has made SCP feasible for the first time. This unlocked applications in cancer biology, immunology, and neuroscience. But proteins are less abundant than nucleic acids and, unlike those, cannot be “amplified”: sample preparation remains the critical bottleneck. Without robust, low-loss methods, sensitivity and reproducibility suffer. Reliable single cell proteomics preparation workflows are therefore essential to fully realize the promise of SCP and to transform protein-level insights into meaningful biological discoveries.
You're in good company
These leading institutions use our single cell and 3D cell model technologies:






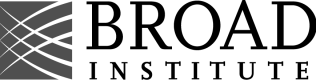



single cell Proteomics applications
Label-free single cell proteomics
Label-free single-cell proteomics directly quantifies proteins from individual cells, with each LC–MS/MS run dedicated to a single lysed and digested cell.
MULTIPLEXED SINGLE CELL PROTEOMICS
Multiplexed single-cell proteomics uses isobaric labeling to combine peptides from many single cells into a pooled LC–MS/MS run. By analyzing dozens of cells simultaneously,, , this approach dramatically increases throughput and sensitivity.
This enables robust comparative studies across large populations. While individual depth is lower than label-free methods, multiplexing is ideal for high-content screens and experiments where reproducibility and statistical power are critical.
SPATIAL PROTEOMICS AT SINGLE-CELL RESOLUTION
Spatial proteomics applies the principles of single-cell proteomics directly within tissues, revealing protein expression in spatial context. Like SCP, it faces bottlenecks of precision, sensitivity, and low input.
By combining accurate isolation with miniaturized workflows, researchers can map proteins across complex tissues while preserving biological context and heterogeneity.
Making Single-Cell Proteomics Workflows Accessible
Whether multiplexed, label-free, or spatial, all single-cell proteomics approaches share the same challenges: ultra-low input, risk of sample loss, and the need for precise, reproducible preparation. cellenONE and proteoCHIP were designed to address these bottlenecks by automating critical steps, minimizing losses, and enabling robust data generation across experimental designs.
Typical Single cell PROTEOMICS workflow
Featured resources

VIDEO
Collaborative Proof-of-Concepts in Single-Cell Proteomics
In this video from Single Cell Proteomics Day, David Hartlmayr presents proof-of-concept results generated with cellenONE® and proteoCHIP® in collaboration with the IMP Vienna Biocenter (Prof. Karl Mechtler’s team), Dr. Claudia Cortecka, and mass spec partners Bruker, Thermo Fisher, and Evosep. Both multiplexed and label-free SCP workflows show improved reproducibility and reduced sample loss thanks to robust, automated preparation.
Publication
Massively Parallel nPOP Sample Preparation for Single-Cell Proteomics, Leduc et al. (Nat Meth) 2024
The nPOP method, developed at Prof Nikolai Slavov’s lab, Northeastern University (Boston, USA), prepares thousands of single cells in nanoliter droplets on glass slides using automated picoliter dispensing with cellenONE®. It underpins multiplexed single-cell proteomics approaches such as plexDIA and isobaric labeling, enabling workflows that process over 1,800 cells per day with proteome coverage of 800–1,200 proteins per cell.
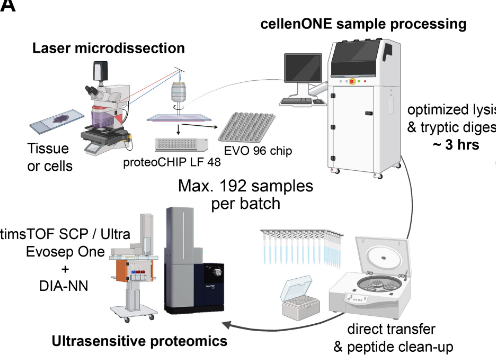
Publication
Automated Spatial Proteomics Workflow with Laser Microdissection, cellenONE and proteoCHIP, Makhmut et al. (MCP) 2024
This study by Prof Fabian Coscia’s group at Max-Delbrück Center (Berlin, GE) presents an automated workflow linking laser microdissection with the cellenONE instrument for ultrasensitive spatial proteomics. Processing up to 192 samples in 3–4 hours on proteoCHIP LF 48 or EVO 96, the protocol minimizes loss, streamlines preparation, and delivers deep proteome coverage (~2,000 proteins) from defined tissue microregions, enabling high-specificity phenotyping in clinical and archival samples.